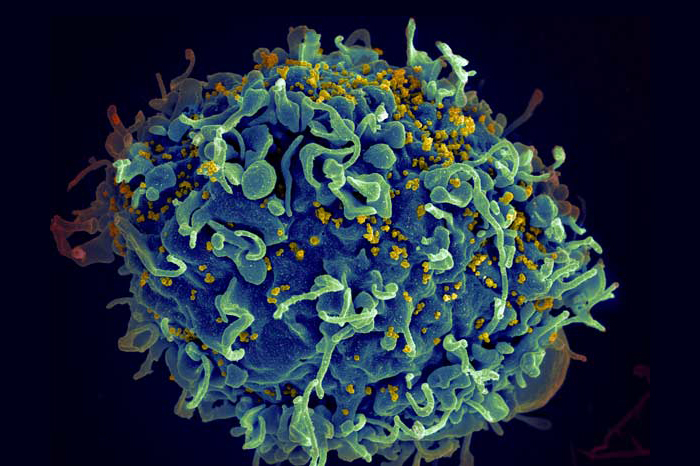
Les ingénieurs en biologie du MIT ont conçu un moyen de réaliser des cribles à grande échelle sur la façon dont les cellules T, comme celle-ci, reconnaissent des agents pathogènes spécifiques, tels que les virus VIH (en jaune) présentés sur cette image. Crédit : Seth Pincus, Elizabeth Fischer et Austin Athman, Institut national des allergies et des maladies infectieuses/NIH.
Les ingénieurs en biologie du MIT ont mis au point un moyen simple d’identifier les cellules B ou T qui interagissent avec des protéines virales ou bactériennes.
Le corps humain compte des millions de cellules B et T uniques qui parcourent l’organisme à la recherche d’envahisseurs microbiens. La capacité de ces cellules immunitaires à reconnaître les microbes nuisibles est essentielle pour combattre efficacement les infections.
MIT biological engineers have now devised an experimental tool that allows them to precisely pick out interactions between a particular immune cell and its target antigen. The new technique, which uses engineered viruses to present many different antigens to huge populations of immune cells, could allow large-scale screens of such interactions.
“This technique leads the way to understand immunity much closer to how the immune system itself actually works, will help researchers make sense of complex immune recognition in a variety of diseases, and could accelerate the development of more effective vaccines and immunotherapies,” says Michael Birnbaum, an associate professor of biological engineering at MIT, a member of MIT’s Koch Institute for Integrative Cancer Research, and the senior author of the study.
Former MIT graduate student Connor Dobson is the lead author of the paper, which was published on April 8th, 2022, in Nature Methods.
A simple screen for a complex system
Both B and T cells play critical roles in launching an immune response. When a T cell encounters its target, it starts proliferating to produce an army of identical cells that can attack infected cells. And B cells that encounter their target begin producing antibodies that help recruit other components of the immune system to clear the infection.
Scientists who study the immune system have several tools to help them identify specific antigen-immune cell interactions. However, these tools generally only allow for the study of a large pool of antigens exposed to one B or T cell, or a large pool of immune cells encountering a small number of antigens.
“In your body, you have millions of unique T cells, and they could recognize billions of possible antigens. All of the tools that have been developed to this point are really designed to look at one side of that diversity at a time,” Birnbaum says.
The MIT team set out to design a new tool that would let them screen huge libraries of both antigens and immune cells at the same time, allowing them to pick out any specific interactions within the vast realm of possibilities.
To create a simple way to screen so many possible interactions, the researchers engineered a specialized form of a lentivirus, a type of virus that scientists often use to deliver genes because it can integrate pieces of DNA into host cells. These viruses have an envelope protein called VSV-G that can bind to receptors on the surface of many types of human cells, including immune cells, and infect them.
For this study, the researchers modified the VSV-G protein so that it cannot infect a cell on its own, instead relying on an antigen of the researchers’ choosing. This modified version of VSV-G can only help the lentivirus get into a cell if the paired antigen binds to a human B or T-cell receptor that recognizes the antigen.
Once the virus enters, it integrates itself into the host cell’s genome. Therefore, by sequencing the genome of all the cells in the sample, the researchers can discover both the antigen expressed by the virus that infected the cell and the sequence of the T or B-cell receptor that allowed it to enter.
“In this way, we can use viral infection itself as a way to match up and then identify antigen-immune cell parings,” Birnbaum says.
Interactions identified
To demonstrate the accuracy of their technique, the researchers created a pool of viruses with antigens from 100 different viruses, including influenza, cytomegalovirus, and Epstein-Barr virus. They screened these viruses against about 400,000 T cells and showed that the technique could correctly pick out antigen-T-cell receptor pairings that had been previously identified.
The researchers also screened two different B-cell receptors against 43 antigens, including antigens from HIV and the spike protein of SARS-CoV-2.
In future studies, Birnbaum hopes to screen thousands of antigens against B and T cell populations. “Our ideal would be to screen entire viruses or families of viruses, to be able to get a readout of your entire immune system in one experiment,” he says.
In one study that is now ongoing, Birnbaum’s lab is working with researchers at the Ragon Institute of MGH, MIT, and Harvard to study how different people’s immune systems respond to viruses such as HIV and SARS-CoV-2. Such studies could help to reveal why some people naturally fight off certain viruses better than others, and potentially lead to the development of more effective vaccines.
The researchers envision that this technology could also have other uses. Birnbaum’s lab is now working on adapting the same viruses to deliver engineered genes to target cells. In that case, the viruses would carry not only a targeting molecule but also a novel gene that would be incorporated exclusively into cells that have the right target. This could offer a way to selectively deliver genes that promote cell death into cancer cells, for example.
“We built this tool to look for antigens, but there’s nothing particularly special about antigens,” Birnbaum says. “You could potentially use it to go into specific cells in order to do gene modifications for cell and gene therapy.”
The research was funded by the Koch Institute Frontier Award program, the Packard Foundation, the Damon Runyon Cancer Research Foundation, the Michelson Medical Research Foundation, Pfizer, Inc., the Department of Defense, the National Institutes of Health, a National Science Foundation Graduate Research Fellowship, a Siebel Scholarship, a Canadian Institutes of Health Research Doctoral Foreign Study Award, a graduate fellowship from the Ludwig Center at MIT, a Medical Scientist Training Program grant from the National Institute of General Medical Sciences, a Technology Impact Award from the Cancer Research Institute, the Pew-Stewart Scholarship program, and the Koch Institute Support (core) Grant from the National Cancer Institute.
Reference: “Antigen identification and high-throughput interaction mapping by reprogramming viral entry” by Connor S. Dobson, Anna N. Reich, Stephanie Gaglione, Blake E. Smith, Ellen J. Kim, Jiayi Dong, Larance Ronsard, Vintus Okonkwo, Daniel Lingwood, Michael Dougan, Stephanie K. Dougan, and Michael E. Birnbaum, 8 April 2022, Nature Methods.
DOI: 10.1038/s41592-022-01436-z


