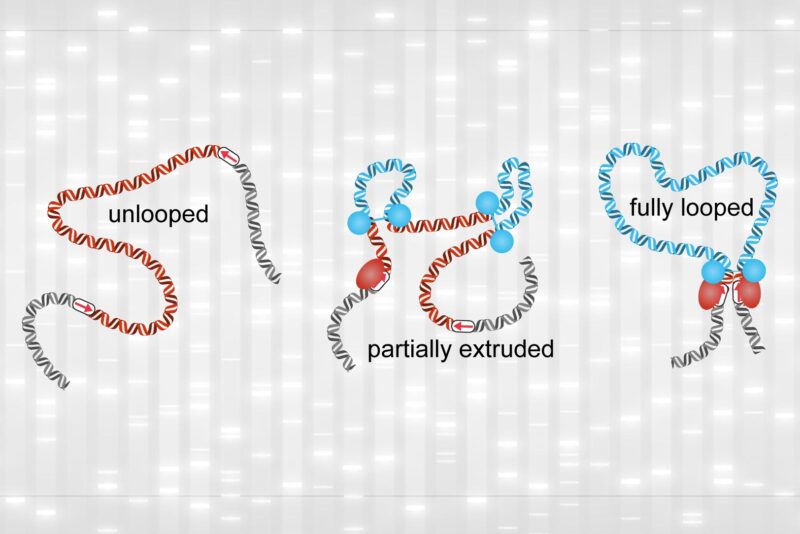
Des chercheurs du MIT ont découvert que la chromatine passe la plupart de son temps dans un état partiellement bouclé (au milieu). Les boucles entièrement formées (à droite) ne se produisent que trois à six pour cent du temps. Crédit : avec l’aimable autorisation des chercheurs, édité par MIT News.
MIT research finds genome loops don’t last long in cells; theories of how loops control gene expression may need to be revised.
In human chromosomes, DNA is coated by proteins to form an extremely long beaded string. This “string” is folded into multiple loops, which are thought to aid cells in controlling gene expression and facilitating DNA repair, among other functions. According to a new MIT study, these loops are more dynamic and shorter-lived than previously thought.
The researchers were able to track the movement of one stretch of the genome in a living cell for roughly two hours in the latest study. They discovered that this segment was only fully looped 3 to 6% of the time, with the loop lasting only about 10 to 30 minutes. According to the researchers, the findings suggest that scientists’ present understanding of how loops regulate gene expression may need to be changed.
“Many models in the field have been these pictures of static loops regulating these processes. What our new paper shows is that this picture is not really correct,” says Anders Sejr Hansen, the Underwood-Prescott Career Development Assistant Professor of Biological Engineering at MIT. “We suggest that the functional state of these domains is much more dynamic.”
Hansen is one of the senior authors of the new study, along with Leonid Mirny, a professor in MIT’s Institute for Medical Engineering and Science and the Department of Physics, and Christoph Zechner, a group leader at the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, Germany, and the Center for Systems Biology Dresden. MIT postdoc Michele Gabriele, recent Harvard University PhD recipient Hugo Brandão, and MIT graduate student Simon Grosse-Holz are the lead authors of the paper, which was published on April 14, 2022, in the journal Science.
Out of the loop
Using computer simulations and experimental data, scientists including Mirny’s group at MIT have shown that loops in the genome are formed by a process called extrusion, in which a molecular motor promotes the growth of progressively larger loops. The motor stops each time it encounters a “stop sign” on DNA. The motor that extrudes such loops is a protein complex called cohesin, while the DNA-bound protein CTCF serves as the stop sign. These cohesin-mediated loops between CTCF sites were seen in previous experiments.
However, those experiments only offered a snapshot of a moment in time, with no information on how the loops change over time. In their new study, the researchers developed techniques that allowed them to fluorescently label CTCF DNA sites so they could image the DNA loops over several hours. They also created a new computational method that can infer the looping events from the imaging data.
“This method was crucial for us to distinguish signal from noise in our experimental data and quantify looping,” Zechner says. “We believe that such approaches will become increasingly important for biology as we continue to push the limits of detection with experiments.”
The researchers used their method to image a stretch of the genome in mouse embryonic stem cells. “If we put our data in the context of one cell division cycle, which lasts about 12 hours, the fully formed loop only actually exists for about 20 to 45 minutes, or about 3 to 6 percent of the time,” Grosse-Holz says.
“If the loop is only present for such a tiny period of the cell cycle and very short-lived, we shouldn’t think of this fully looped state as being the primary regulator of gene expression,” Hansen says. “We think we need new models for how the 3D structure of the genome regulates gene expression, DNA repair, and other functional downstream processes.”
While fully formed loops were rare, the researchers found that partially extruded loops were present about 92 percent of the time. These smaller loops have been difficult to observe with the previous methods of detecting loops in the genome.
“In this study, by integrating our experimental data with polymer simulations, we have now been able to quantify the relative extents of the unlooped, partially extruded, and fully looped states,” Brandão says.
“Since these interactions are very short, but very frequent, the previous methodologies were not able to fully capture their dynamics,” Gabriele adds. “With our new technique, we can start to resolve transitions between fully looped and unlooped states.”
Les chercheurs émettent l’hypothèse que ces boucles partielles pourraient jouer un rôle plus important dans la régulation des gènes que les boucles entièrement formées. Les brins d’ADN se déplacent les uns le long des autres lorsque les boucles commencent à se former puis se désagrègent, et ces interactions peuvent aider les éléments régulateurs tels que les exhausteurs et les promoteurs de gènes à se trouver les uns les autres.
“Plus de 90 % du temps, il y a des boucles transitoires, et ce qui est probablement important, c’est d’avoir ces boucles qui sont perpétuellement extrudées”, dit Mirny. “Le processus d’extrusion lui-même est peut-être plus important que l’état de boucle complète qui ne se produit que pendant une courte période de temps.”
D’autres boucles à étudier
Puisque la plupart des autres boucles du génome sont plus faibles que celle que les chercheurs ont étudiée dans cet article, ils soupçonnent que de nombreuses autres boucles se révéleront également très transitoires. Ils prévoient maintenant d’utiliser leur nouvelle technique pour étudier certaines de ces autres boucles, dans une variété de types de cellules.
“Il existe environ 10 000 de ces boucles, et nous n’en avons étudié qu’une seule”, déclare Hansen. “Nous avons beaucoup de preuves indirectes pour suggérer que les résultats seraient généralisables, mais nous ne l’avons pas démontré. En utilisant la plateforme technologique que nous avons mise en place, qui combine de nouvelles méthodes expérimentales et informatiques, nous pouvons commencer à aborder d’autres boucles du génome.”
Les chercheurs prévoient également d’étudier le rôle de boucles spécifiques dans les maladies. De nombreuses maladies, dont un trouble neurodéveloppemental appelé syndrome FOXG1, pourraient être liées à une dynamique de boucle défectueuse. Les chercheurs étudient actuellement comment la forme normale et mutée du gène FOXG1, ainsi que le gène cancérigène MYC, sont affectés par la formation de boucles génomiques.
Référence : “Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging” par Michele Gabriele, Hugo B. Brandão, Simon Grosse-Holz, Asmita Jha, Gina M. Dailey, Claudia Cattoglio, Tsung-Han S. Hsieh, Leonid Mirny, Christoph Zechner et Anders S. Hansen, 14 avril 2022, Science.
DOI : 10.1126/science.abn6583
La recherche a été financée par les National Institutes of Health, la National Science Foundation, la Mathers Foundation, une bourse Pew-Stewart Cancer Research Scholar, les Chaires d’excellence Internationale Blaise Pascal, une bourse de recherche American-Italian Cancer Foundation, et l’Institut Max Planck de biologie cellulaire moléculaire et de génétique.