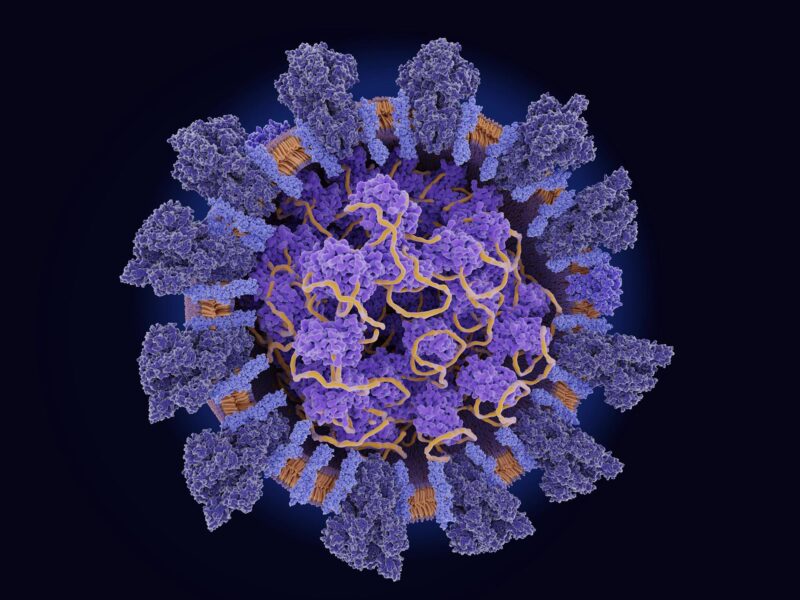
Structure du coronavirus SRAS-CoV-2.
La connaissance de la fonction de la protéine de la nucléocapside pourrait aider à développer des médicaments qui réduisent l’impact du coronavirus.
Une collaboration multicentrique qui suit la propagation et l’évolution du SARS-CoV-2 virus in Saudi Arabia has identified mutations in the virus’s N protein associated with increased viral loads in COVID-19 patients. The study provides insight into the function of this nucleocapsid protein, which could help develop drugs that reduce the impact of coronavirus infection.
“The nucleocapsid (N) protein is the most abundant protein in all coronaviruses, including SARS-CoV-2,” explains KAUST research scientist Muhammad Shuaib. This protein binds to various parts of the viral RNA, affecting how it is packaged within the virus. It also plays roles inside host cells related to viral replication and host immune responses.
The researchers, working with Arnab Pain, found that two consecutive mutations in the N protein, called R203K and G204R, were associated with increased severity of COVID-19 in patients. Analyses showed that the changes to the protein made it bind more strongly to the viral RNA.

From left: Sharif M. Hala, Sara Mfarrej, Professor Arnab Pain, Muhammad Shuaib and Tobias Mourier (not pictured) studied the N protein of SARS-CoV-2 to further understanding of its role in disease severity. Credit: © 2022 KAUST
Further tests in laboratory cells suggested that the changes in the N protein allow the virus to more efficiently hijack host cell translation machinery to facilitate virus replication. They were also associated with increased expression of genes involved in interferon and chemokine production. This could be behind the life-threatening cytokine storm that happens in some COVID-19 patients, making it very difficult for them to breathe.
The findings were the result of analyses of viral genome sequences from 892 patient samples taken from various parts of Saudi Arabia between March and August 2020, relatively early in the pandemic. This was followed by comparisons with patient data to understand how mutations affected viral load and virulence.
“Compared to the spike protein, the N protein is highly conserved in the different coronaviruses, like SARS and MERS; yet attempts to design vaccines against it have not been successful,” says Tobias Mourier, a consultant research scientist working on Pain’s team. “Understanding N protein’s function could help develop drugs that target it and potentially limit disease severity in COVID-19 and other coronavirus infections.”
The KAUST-led research team, which includes scientists and clinicians from institutions and hospitals all over Saudi Arabia, continues to monitor the SARS-CoV-2 virus nationwide and observe how mutations affect virus-host interactions under various vaccination regimens. “Sequencing virus genomes and reporting genomic changes from regions of the world that are severely underrepresented in current databases is essential for tracking and assessing new variants of concern,” says Pain.
Reference: “SARS-CoV-2 genomes from Saudi Arabia implicate nucleocapsid mutations in host response and increased viral load” by Tobias Mourier, Muhammad Shuaib, Sharif Hala, Sara Mfarrej, Fadwa Alofi, Raeece Naeem, Afrah Alsomali, David Jorgensen, Amit Kumar Subudhi, Fathia Ben Rached, Qingtian Guan, Rahul P. Salunke, Amanda Ooi, Luke Esau, Olga Douvropoulou, Raushan Nugmanova, Sadhasivam Perumal, Huoming Zhang, Issaac Rajan, Awad Al-Omari, Samer Salih, Abbas Shamsan, Abbas Al Mutair, Jumana Taha, Abdulaziz Alahmadi, Nashwa Khotani, Abdelrahman Alhamss, Ahmed Mahmoud, Khaled Alquthami, Abdullah Dageeg, Asim Khogeer, Anwar M. Hashem, Paula Moraga, Eric Volz, Naif Almontashiri and Arnab Pain, 1 February 2022, Nature Communications.
DOI: 10.1038/s41467-022-28287-8